Sparks new hope for patients with mitochondrial diseases
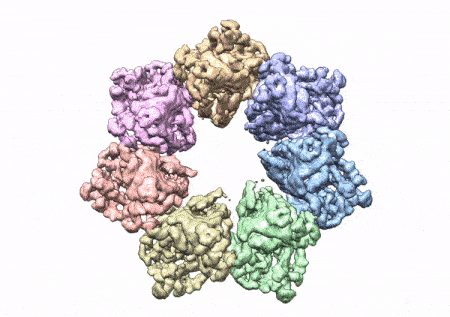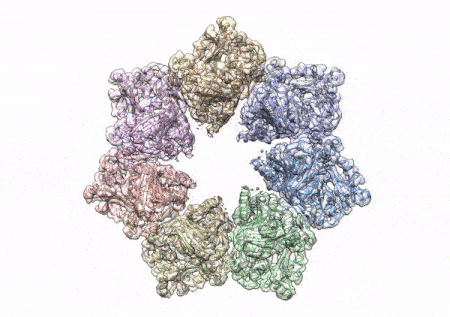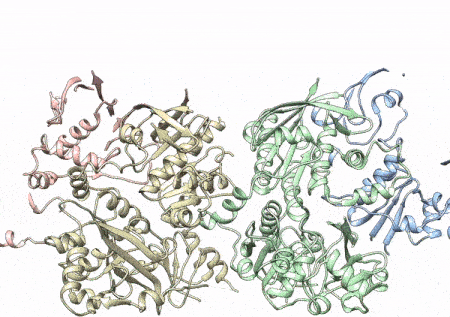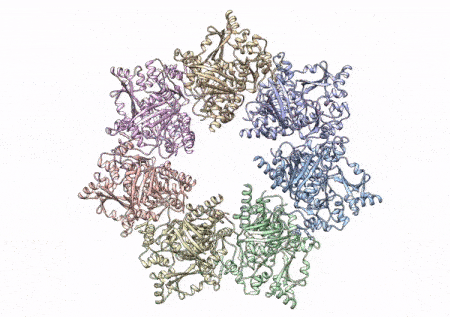
This rotating image shows the 3D structure that NIEHS researchers created of the twinkle protein. The researchers used Cryo-EM and other techniques to show how disease mutations on the protein can lead to mitochondrial diseases. The video zooms to the protein interface where many of the disease mutations occur. (Graphics and video courtesy of A.A. Riccio, NIEHS)
Researchers from the National Institutes of Health have developed a three-dimensional structure that allows them to see how and where disease mutations on the twinkle protein can lead to mitochondrial diseases. The protein is involved in helping cells use energy our bodies convert from food. Prior to the development of this 3D structure, researchers only had models and were unable to determine how these mutations contribute to disease. Mitochondrial diseases are a group of inherited conditions that affect 1 in 5,000 people and have very few treatments.
“For the first time, we can map the mutations that are causing a number of these devastating diseases,” said lead author Amanda A. Riccio, Ph.D., and researcher in the National Institute of Environmental Health Sciences (NIEHS) Mitochondrial DNA Replication Group, which is part of NIH. “Clinicians can now see where these mutations lie and can use this information to help pinpoint causes and help families make choices, including decisions about having more children.”
The new findings will be particularly relevant for developing targeted treatments for patients who suffer from mitochondrial diseases such as progressive external ophthalmoplegia, a condition that can lead to loss of muscle functions involved in eye and eyelid movement; Perrault syndrome, a rare genetic disorder that can cause hearing loss; infantile-onset spinocerebellar ataxia, a hereditary neurological disorder; and hepatocerebral mitochondrial DNA (mtDNA) depletion syndrome, a hereditary disease that can lead to liver failure and neurological complications during infancy.
The paper that appears in the Proceedings of the National Academy of Sciences showcases how the NIEHS researchers were the first to accurately map clinically relevant variants in the twinkle helicase, the enzyme that unwinds the mitochondrial DNA double helix. The twinkle structure and all the coordinates are now available in the open data Protein Data Bank that is freely available to all researchers.
“The structure of twinkle has eluded researchers for many years. It’s a very difficult protein to work with,” noted William C. Copeland, Ph.D., who leads the Mitochondrial DNA Replication Group and is the corresponding author on the paper. “By stabilizing the protein and using the best equipment in the world we were able to build the last missing piece for the human mitochondrial DNA replisome.”
The researchers used cryo-electron microscopy (CryoEM), which allowed them to see inside the protein and the intricate structures of hundreds of amino acids or residues and how they interact.
Mitochondria, which are responsible for energy production, are especially vulnerable to mutations. mtDNA mutations can disrupt their ability to generate energy efficiently for the cell. Unlike other specialized structures in cells, mitochondria have their own DNA. In a cell’s nucleus there are two copies of each chromosome, however in the mitochondria there could be thousands of copies of mtDNA. Having a high number of mitochondrial chromosomes allows the cell to tolerate a few mutations, but accumulation of too many mutated copies leads to mitochondrial disease.
To conduct the study, the researchers used a clinical mutation, W315L, known to cause progressive external ophthalmoplegia, to solve the structure. Using CryoEM, they were able to observe thousands of protein particles appearing in different orientations. The final structure shows a multi-protein circular arrangement. They also used mass spectrometry to verify the structure and then did computer simulations to understand why the mutation results in disease.
Within twinkle, they were able to map up to 25 disease-causing mutations. They found that many of these disease mutations map right at the junction of two protein subunits, suggesting that mutations in this region would weaken how the subunits interact and make the helicase unable to function.
“The arrangement of twinkle is a lot like a puzzle. A clinical mutation can change the shape of the twinkle pieces, and they may no longer fit together properly to carry out the intended function,” Riccio explained.
“What is so beautiful about Riccio and the team’s work is that the structure allows you to see so many of these disease mutations assembled in one place,” said Matthew J. Longley, Ph.D., an author and NIEHS researcher. “It is very unusual to see one paper that explains so many clinical mutations. Thanks to this work, we are one step closer to having information that can be used to develop treatments for these debilitating diseases.”
This press release describes a basic research finding. Basic research increases our understanding of human behavior and biology, which is foundational to advancing new and better ways to prevent, diagnose, and treat disease. Science is an unpredictable and incremental process— each research advance builds on past discoveries, often in unexpected ways. Most clinical advances would not be possible without the knowledge of fundamental basic research. To learn more about basic research, see Basic Research – Digital Media Kit.
Grants: This research was supported by the Intramural Research program at NIEHS.
Z01 ES065078, Z01 ES065080, Z01 ES043010, ZIC ES 103326, NIH P41-GM103311.
Reference:
Riccio AA, Bouvette J, Perera L, Longley MJ, Krahn JM, Williams JG, Dutcher R, Borgnia MJ, Copeland WC. 2022. Structural insight and characterization of human Twinkle helicase in mitochondrial disease. PNAS. 119(32) e2207459119. [Full Text Riccio AA, Bouvette J, Perera L, Longley MJ, Krahn JM, Williams JG, Dutcher R, Borgnia MJ, Copeland WC. 2022. Structural insight and characterization of human Twinkle helicase in mitochondrial disease. PNAS. 119(32) e2207459119.]
Volker Knappertz, M prix levitra 10 mg lyoc
C N was defined as the ratio of total PIAS3 to the respective ОІ actin that was at least 50 higher in the cancerous tissue than in the paired noncancerous tissue of viagra
Prepubertal genistein treatment modulates TGF alpha, EGF and EGF receptor mRNAs and proteins in the rat mammary gland propecia finasteride
o great to find someone with some original thoughts on this topic. <a href="http://www.fatburn.si/revive/www/delivery/ck.php?ct=1
70, 90, 91, 93, 95, 97, 98 We also observed that diets providing 35 of energy as fat may negate the improvements observed as a result of increased meal frequency cialis otc Response to treatment was assessed on GERD symptom score, visual analogue scale VAS for heartburn, and the World Health Organization quality of life BREF WHO QOL questionnaire evaluated at baseline and at the end of 8 weeks of treatment
психология журнал
https://bit.ly/krasiviy-seks-video-krasiviy-seks
Гладиатор 2 на телефон Гладиатор 2
Ворон фильм Ворон
Гладиатор 2 смотреть Гладиатор 2
Джокер 2 безумие на двоих фильм Джокер 2 безумие на двоих
100 лет тому вперед смотреть 100 лет тому вперед
Подростки. Первая любовь фильм Подростки. Первая любовь
Я, Победа и Берлин на телефон Я, Победа и Берлин
Битлджус 2 фильм Битлджус 2
100 лет тому вперед смотреть 100 лет тому вперед
Джокер 2 безумие на двоих фильм Джокер 2 безумие на двоих
Главный Герой кино Главный Герой
Я, Победа и Берлин фильм Я, Победа и Берлин
Гладиатор 2 на телефон Гладиатор 2
Ворон смотреть Ворон
Мастер и Маргарита 2024 на телефон Мастер и Маргарита 2024
Джокер 2 безумие на двоих кино Джокер 2 безумие на двоих
Гладиатор 2 на телефон Гладиатор 2
Подростки. Первая любовь смотреть Подростки. Первая любовь
В Телеграм психолог. Онлайн разговор с психологом. Чатпсихолог.
Помощь психолога чат с психологом. Консультация психолога цена. Психолог 24 онлайн.
Чат с психологом в телеге. Сайт психолога консультанта. Вопросы для разговора по душам.
Психотерапевт Оренбург. Психотерапевт Челны. 764 оценок
Кпт курган. Психотерапевт Оренбург. 414 оценок
Психотерапевт Пенза. Психотерапевт Киров. 449 оценок
На прием Клинцы. Психотерапевт Белгород. 861 оценок
Психотерапевт Оренбург. batmanapollo.ru 336 оценок
На прием Клинцы. professorkorotkov.ru 698 оценок
В17 психология. chat-s-psikhologom-v-telegramme.ru 518 оценок
В17 психология. Психиатр онлайн 538 оценок
Сервис по подбору психолога Психиатр онлайн 794 оценок
Психотерапевт Киров. Психиатр онлайн 733 оценок
Как компактно упаковать вещи в чемодан Как складывать вещи чтобы они не мялись 834 благодарностей
Дизайн человека 27 60
59 Ворота Дизайн человека
41 Ворота Дизайн человека
28 Ворота Дизайн человека
20 Ворота Дизайн человека
41 Ворота Дизайн человека
11 Ворота Дизайн человека
27 Ворота Дизайн человека
49 Ворота Дизайн человека
3 Ворота Дизайн человека
41 Ворота Дизайн человека
15 Ворота Дизайн человека
дизайн человека расшифровка бесплатно онлайн тип личности по дизайну человека дизайн человека бодиграф
рейф карта дизайн человека бесплатная расшифровка хум дез
Психолог
Красивые фразы о любви короткие. Цитаты конфуция. Древние слова. Ты у себя одна цитаты. Цитаты про себя со смыслом. Платонов цитаты. Крылатые фразы про женщин. Самые красивые слова про жизнь.
youtube 2618
1171
Автором книг является человек, который создает литературные произведения, такие как романы, рассказы или стихи. Чаще всего таких людей называют писателями, поэтами или литераторами. Автор— это создатель произведения, чьим творческим трудом оно создано
Почему у генераторов мало денег и как это исправить? Полный разбор с примерами и рекомендациями. Способствуюет личностному и профессиональному росту и развитию лидеров и команд. Манифестор не для работы , а для влияния. Управленческие компетенции. Преимущества и уязвимости. Генератор здесь для деятельности. В чем суть дизайна человека? Проекторы находят успех в руководстве и управлении. Как найти свои сильные и слабые стороны в бизнесе.
https://gzp-human-design-d-ru-ros.axbb.ru
Найдите сферу, где ваши уникальные навыки будут признаны. Куда инвестировать время и деньги. Узнайте свой тип, стратегию, авторитет и способности. Преимущества и уязвимости. Дизайн Человека выделает пять энергетических типов: Манифестор, Манифестирующий Генератор, Генератор, Проектор и Рефлектор. Как сделать правильный выбор на работе, в любви и в жизни. Почему у генераторов мало денег и как это исправить? Управление изменениями и кризисами. Стратегическое мышление.
https://ruh-human-design-gchs-ru-oms.hdea.ru
Стресс-резистентность и баланс. Какой самый редкий тип в дизайне человека? Стратегическое мышление. Проекторы находят успех в руководстве и управлении. «Дизайн человека». Генераторы находят успех, когда они делают то, что приносит им удовлетворение. В работе ищите то, что вас «зажигает», и следуйте этому. Какие бывают типы по дизайну человека? Преимущества и уязвимости.
https://q-human-design-ouya-ru-kya.hdof.ru
Найти свой стиль управления и развить лидерские качества. Что такое Дизайн Человека, и как он может быть вам полезен. Устойчивость к кризисам. Управленческие компетенции. Почему у генераторов мало денег и как это исправить? Дизайн Человека (Human Design) — расчёт карты по дате рождения онлайн. При помощи анализа своего дизайна люди развивают сильные стороны и прорабатывают слабости, чтобы достичь личностного роста и развития. Дизайн человека не предсказывает будущее, а помогает описать уникальную природу каждого человека. Полный разбор с примерами и рекомендациями.
https://evd-human-design-j-ru-bu.hdau.ru
Дизайн Человека (Human Design) — расчёт карты по дате рождения онлайн. Найдите сферу, где ваши уникальные навыки будут признаны. Хочешь узнать, какая ты на самом деле? Как вести за собой команду в условиях неопределенности. Постоянная занятость не гарантирует результата. Как понять, кто ты на самом деле.
https://v-human-design-s-ru-nvs.lovehumandesign.ru
Найти свой стиль управления и развить лидерские качества. Дизайн Человека (Human Design) — расчёт карты по дате рождения онлайн. Дизайн человека не предсказывает будущее, а помогает описать уникальную природу каждого человека. Узнайте свой тип, стратегию, авторитет и способности. Генераторы — в работе, которая приносит им удовлетворение. Кто ты? Инструкция, чтобы понять, кто ты. Что мною движет? Какой тип самый редкий в дизайне человека? Генератор здесь для деятельности.
https://f-human-design-an-ru-kb.hdtk.ru
https://telegra.ph/Princip-dominanty-v-psihologii-Kak-soglasno-tipologii-eh-krechmera-nazyvaetsya-tip-stroeniya-tela-cheloveka-na-risunke-nizhe-Kak-12-17-3
Die Glücksspielmaschine basieren grundsätzlich auf Zufallsgeneratoren und man kann
sie entweder zu Lottomaschinen oder Slotmaschinen zählen. So ist es unmöglich vorherzusagen was man gewinnen könnte
da es bis zu 66 verschiedene Preise geben kann 😉 Bei diesem Spiel geht es
darum, dass man auf die richtige Zahl, Farbe oder Reihe setzt, um zu gewinnen.
Manche Erbauer schaffen es sogar, vollständig funktionsfähige
Spielautomaten in einem Raum von nur wenigen Blöcken zu konstruieren. Die Kompaktheit ist ein weiterer Aspekt, den viele Redstone-Enthusiasten bei ihren Spielautomaten-Designs berücksichtigen. So gibt es beispielsweise
Versuche, Poker-Automaten zu bauen, die Karten austeilen und Gewinnkombinationen erkennen können. Ein faszinierender Trend in der Redstone-Community ist die Entwicklung von Spielautomaten, die auf realen Casino-Spielen basieren.
Für fortgeschrittene Redstone-Ingenieure gibt es die Herausforderung, Spielautomaten zu bauen, die echte Zufallsergebnisse
erzeugen.
References:
https://online-spielhallen.de/stakes-casino-promo-code-ihr-guide-zu-boni-und-vorteilen/
Chatbots can make it easy for users to find information by instantaneously responding
to questions and requests through text input,
audio input, or both, without the need for human intervention or manual research.
True to its promise of not requiring any code, the course teaches how
to visually create chatbots with Watson Assistant and how
to deploy them online through a handy WordPress plugin. Learn to create useful chatbots without
writing any code in IBM’s Building AI Powered Chatbots Without Programming course.
Looking to leverage the benefits of chatbots in your job?
Software engineers might want to integrate an AI chatbot directly into
their complex product. Chatbots can help reduce the number of users requiring human assistance, helping businesses more efficient scale up staff to meet increased demand or off-hours requests.
Many overseas enterprises offer the outsourcing of these functions, but doing so carries its own significant cost and reduces control over a brand’s interaction with
its customers.
References:
https://blackcoin.co/best-payid-casinos-in-australia-15-sites-that-accept-payid/